An increased understanding of the genomic and molecular underpinnings of cancer pathogenesis and their exploitation in the development of targeted therapies has revolutionized the treatment of many cancer types including BRAF-mutant melanoma, HER2-amplified breast cancer, and non–small cell lung cancer (NSCLC) harboring epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase rearrangements. Identification of biomarkers has been a prerequisite for the success of these targeted therapies to confirm target inhibition, guide patient selection, and understand mechanisms of resistance.
More recently, immunologic therapy has emerged as an important treatment option for many types of cancers, based on demonstrations of unprecedented efficacy. This radical shift in treatment has come with the recognition of the essential role of the immune system in the surveillance and eradication of neoplastic cells, particularly modulation of the immune checkpoint protein cytotoxic T-lymphocyte–associated antigen 4 (CTLA-4) and the programmed death-1 (PD-1) receptor and its ligand, PD-L1. The new class of immune checkpoint inhibitors has dramatically changed the treatment landscape of metastatic melanoma, NSCLC, and other tumor types. Despite this remarkable success, other goals remain elusive, such as determining which patients will respond to cancer immunotherapy, experience immune-related adverse events (irAEs), or develop resistance to immunotherapy. Hence, there is a need to identify biomarkers that will provide predictive and prognostic information as well as insights into the mechanisms underlying patient responses or immune escape. This review describes challenges associated with identification of immune biomarkers, testing methodologies, and promising biomarkers that have been assessed thus far in patients treated with immune checkpoint inhibitors.
Clinical Use of Biomarkers
Underscoring the need for a harmonization of terms that guide research into this area and the subsequent development of new therapies, the US Food and Drug Administration (FDA) and the National Institutes of Health jointly sponsored the Biomarkers, EndpointS, and other Tools (BEST) Resource in 2015. There, the term biomarker is identified in part as “[a] defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions.”1 Simplistically, it refers to a measurement variable that is associated with disease outcome.2 The measurement variable may be a molecular, histologic, radiographic, or physiologic characteristic, or a composite of molecular alterations such as gene expression or proteomic signatures.3 Examples of biomarkers include expression of EGFR, prostate-specific antigen (PSA) levels, or the Oncotype DX recurrence score, which is calculated from the measurements of the expression levels of 21 genes.2 For clinical use, biomarker assessments are expected to fulfill certain criteria such as being routine, simple, rapid, robust, cost-effective, reproducible, specific, quantitative, and standardized.4 To be applied in clinical practice, a candidate biomarker must demonstrate analytical validity, clinical validity and utility, and be prospectively validated in randomized clinical trials.3,5
Biomarkers may be used in multiple clinical settings in the continuum of cancer diagnosis, treatment, and follow-up. This includes risk assessment, screening, differential diagnosis, determination of prognosis, response to treatment, disease status, and safety monitoring. In addition, biomarkers may help detect cancer recurrence or progression.3
Biomarkers may also help determine a patient’s risk for developing cancer, allowing them to seek risk-reduction strategies. For example, presence of germline BRCA1 mutation increases a woman’s risk for developing breast and/or ovarian cancer, which may be mitigated with prophylactic mastectomy. In addition, biomarkers may be used for screening purposes in otherwise healthy patients for malignancy (eg, PSA).3
Perhaps most important, biomarkers may be used in the cancer treatment setting with the primary goal of individualizing treatment: to identify optimal therapy, monitor for safety, determine whether a patient will respond to a particular treatment, and to assess overall patient outcomes.3 Three common types of biomarkers most pertinent to clinical practice, as well as basic and clinical research (including clinical trial design and enrollment), are prognostic biomarkers, predictive biomarkers, and pharmacodynamic biomarkers.
A prognostic biomarker provides information regarding the disease course and the likelihood of some clinical outcome, such as disease progression, disease recurrence, or death, independent of the therapy received. For example, the stage of disease based on tumor invasion has traditionally been used as a prognostic biomarker.3 More recently, molecular and genetic tumor characteristics are being used to determine prognosis; for example, PIK3CA mutation status had prognostic value in women with HER2-positive metastatic breast cancer, independent of treatment received.6 A prognostic biomarker may reflect the disease’s underlying biology and natural history in the absence of any treatment, as exemplified by untreated patients with HER2-positive breast cancer showing worse survival compared with untreated patients with HER2-negative disease.7
By contrast, predictive biomarkers may inform clinicians about treatment effect and whether patients are more likely to respond to any given treatment.2 For example, expression of a specific protein on tumor cells directly correlates with the benefit derived from therapies targeting that protein, such as in patients with early-stage HER2-positive breast cancer deriving benefit from HER2-directed therapy.8
Pharmacodynamic biomarkers provide insight into the pharmacologic effects of a drug on its target, including proof of mechanism (ie, does the treatment hit its intended target?), proof of concept (ie, does the drug induce the intended biologic outcome in its target?), selection of optimal dosing/avoidance of toxicity, and understanding of response/resistance mechanisms.9 For example, comparison of serial tumor biopsies performed before, during, and after treatment may reveal reduction in expression of target protein, demonstrating proof of mechanism or acquisition of new resistance mutations.
Biomarkers may also be used to predict adverse reactions associated with a treatment. For example, in patients treated with standard doses of irinotecan, presence of germline genetic mutations (ie, UGT1A1*28 mutation) increased the risk for developing severe neutropenia and diarrhea.3,10 Emerging evidence has documented immune changes preceding irAEs, such as clonal expansion of CD8 T-cells prior to the development of ipilimumab-induced toxicities, or B-cell changes that preceded irAEs following combination immune checkpoint blockade.11,12
In addition, biomarkers may be used to detect early recurrence of disease prior to manifestation of symptoms or to monitor treatment response. For example, in patients with colon cancer, the clinical utility of monitoring carcinoembryonic antigen levels following adjuvant treatment to detect liver metastases has been demonstrated.13
In the drug development process, biomarkers are used in clinical trials in many of the settings described above—such as to demonstrate mechanism of action, inform patient selection, monitor treatment safety and response, and to optimize clinical trial design by stratifying patients based on their biomarker profiles. Importantly, biomarker testing or molecular profiling can improve screening for clinical trials and enhance clinical trial accrual.
Immunologic Biomarkers
There is an unmet need for validated biomarkers specific to immunotherapies that would guide treatment decisions specific to each patient, by avoiding selection of ineffective therapies and toxicities and preventing mechanisms of immune escape. It has become increasingly apparent that the principles used for the development of biomarkers for traditional targeted therapy may not apply to cancer immunotherapy. The primary hurdle limiting research efforts to identify biomarkers is the complexity and dynamic nature of the interactions between the immune system and tumor cells. Therefore, understanding the complex pathobiology of cancer–immune system interactions is paramount.5
The tumor microenvironment (TME) is composed of many cell types, including the heterogeneous tumor cell of origin, fibroblasts, endothelial cells, and a variety of tumor-infiltrating immune cells such as T- and B-lymphocytes, monocytes, neutrophils, dendritic cells, and natural killer cells, as well as tumor-associated macrophages. Within the TME, distinct “microniches” with their own mini-microenvironment may also be present with a different cellular and molecular milieu.14 Activated T-cells are primary mediators of immune effector functions, involving a T-cell response induced by recognition of tumor-specific antigens presented on major histocompatibility complex (MHC) molecules on the cell, followed by initiation of proliferation and functional differentiation of T-cells. The tumor antigens are derived from several sources, including neoantigens from mutations associated with the process of carcinogenesis. Regulation of T-cell responses is a complex process consisting of both stimulatory and inhibitory cell signaling pathways, impacting the overall antitumor response.15
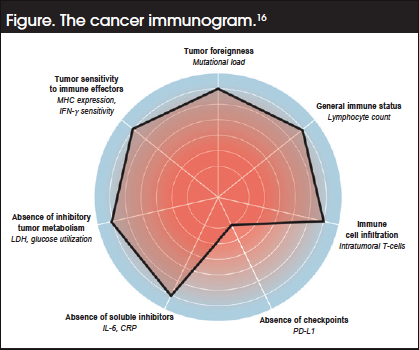
Assuming a predominant role for enhanced T-cell activity in the anticancer immune response, a cancer immunogram has been proposed around a framework of 7 parameter classes, for which biomarkers can potentially be identified (Figure). These parameters are16:
- Tumor foreignness as determined by tumor mutational burden (TMB)
- The general immune status of the patient assessed from lymphocyte counts
- Tumor infiltration of tumor-reactive T-cells
- Expression of PD-L1
- Absence of soluble inhibitors (C-reactive protein and interleukin-6)
- Absence of inhibitory tumor metabolism, with high serum lactate dehydrogenase reflecting both tumor burden and anaerobic glycolysis
- Tumor sensitivity to immune effectors, including MHC expression and the functional interferon-γ receptor pathway
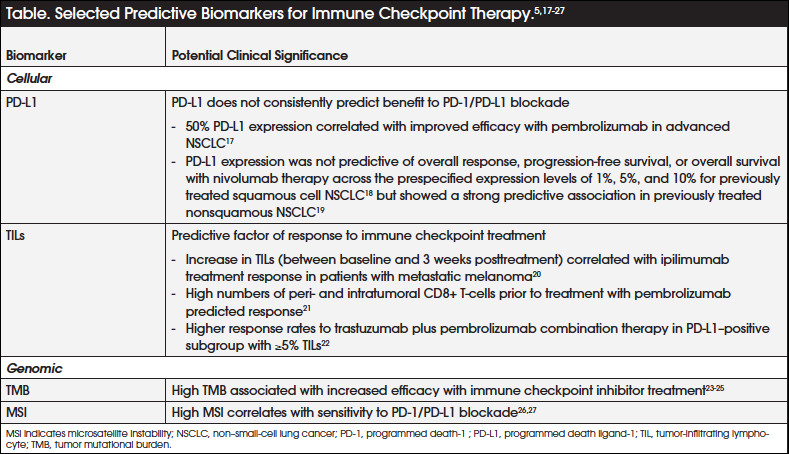
Based on this increased understanding of the tumor–immune system interplay, candidate biomarkers of particular interest are tumor PD-L1 expression, tumor-infiltrating lymphocytes (TILs), TMB, and microsatellite instability (MSI), among others (Table).5,17-27 Of these, the best-studied predictive biomarker of checkpoint inhibitor therapy to date is tumor expression of PD-L1. However, on the basis of available evidence, PD-L1 does not appear to consistently predict benefit of PD-1/PD-L1 blockade.5,28 Although a direct correlation exists between higher tumor PD-L1 expression and favorable outcomes with PD-1/PD-L1 blockade, patients with tumors that do not express PD-L1 may also derive benefit from anti–PD-1 immune checkpoint inhibitors.5,28,29 It is also unclear whether PD-L1 expression on the tumor cells themselves or on immune cells such as lymphocytes and macrophages is most relevant, since higher numbers of PD-1– and PD-L1–expressing tumor-infiltrating CD8+ T-cells correlated with clinical responses in patients with metastatic melanoma treated with the anti–PD-1 inhibitor pembrolizumab.21
Additional challenges to using PD-L1 expression as a biomarker are that its expression is heterogeneous and dynamic within an individual, may be heterogeneous within a tumor sample, and may be discordant between the primary lesion and its metastases, thus not reflecting the true PD-L1 expression status of the tumor.5 For these reasons, PD-L1 is considered an imperfect predictive biomarker for response to immune checkpoint therapy.
Current evidence indicates that foreignness or immunogenicity of a tumor may largely be determined by the expression of neoantigens, which directly correlates with the mutational load of the tumor and the outcome of immune checkpoint therapy.15,30 The mutational load of different tumor types varies, with tumors with high overall mutational load showing a higher likelihood of responding to immune checkpoint therapy.15 This correlation between mutational load and outcome with immune checkpoint inhibitors has been demonstrated in melanoma and NSCLC.23,24
Moreover, a recent report of the results from the CheckMate-227 study demonstrated superior progression-free survival (PFS) with the combination of 2 immune checkpoint inhibitors (nivolumab and ipilimumab) versus chemotherapy as first-line treatment in patients with NSCLC and high TMB, regardless of PD-L1 expression.25 Similarly, pembrolizumab was found to be particularly efficacious in patients with colorectal cancer (CRC) that demonstrated a deficiency in DNA mismatch repair (MMR).27
Surrogate measures of TMB, including defects in DNA repair mechanisms such as MSI, have emerged as clinically useful biomarkers that are predictive of response to immune checkpoint inhibitor treatment.31,32 Although anti–PD-1 therapy was initially thought not to be effective in CRC, further analysis indicates that a subset of cancers characterized by deficient MMR (dMMR) and/or MSI-high (MSI-H) profiles may be particularly sensitive to PD-1/PD-L1 blockade, which is attributed to higher PD-L1/PD-L1 expression.26
Presence of TILs is a favorable prognostic factor as well as a predictive factor of response to immune checkpoint treatment in many types of cancer.30 In patients with metastatic melanoma, clinical responses to ipilimumab correlated with an increase in TILs between baseline and 3 weeks posttreatment; similarly, clinical response to pembrolizumab was shown to correlate with patients having high numbers of peri- and intratumoral CD8+ T-cells prior to pembrolizumab treatment.20,21 Moreover, expression of granzyme B, reflecting CD8+T-cell effector function, was increased after immune checkpoint inhibitor therapy among patients who responded.21
Biomarker Testing Methodologies
Immunohistochemistry (IHC), polymerase chain reaction (PCR), and fluorescence in situ hybridization (FISH) are routinely used for detection of traditional biomarkers. Using target-specific antibodies coupled with colorimetric or fluorescent probes, IHC detects specific antigens and facilitates in situ anatomical localization of proteins. On the other hand, FISH uses fluorescent marker-labeled DNA probes that hybridize with target gene sequences, allowing visualization through a fluorescence microscope.33 PCR is a common technique that is used to amplify specific DNA sequences using short DNA fragments or primers that are complementary to the target DNA.34
Although the availability of novel and powerful high-throughput technologies has led to a significant advancement in the biomarker-based research field in general, these tools are particularly critical for the development of biomarkers for cancer immunotherapies, given the complexity, heterogeneity, and dynamic nature of the cancer–immune system interactions. Moreover, since TILs are heterogeneous and represent only a fraction of the total tumor mass, technologies that can detect low abundance transcripts are needed. Based on these considerations, new biomarker assays, including quantitative real-time PCR-assisted cell counting, protein microarray (seromics), flow and mass cytometry, multiplexed tissue biomarker imaging, and whole exome sequencing have been developed and may pave the way for defining the repertoire of the immune response to cancer.35,36
Next-generation sequencing (NGS) coupled with bioinformatics provides biologic information on a genomewide scale, to accurately identify the immune status of tumors from properly collected and processed specimens.36 NGS uses 2 approaches: the PCR platform or hybrid capture, with the former using multiplex primer pairs to form a library that is suitable for sequencing. Hybrid capture involves hybridization of DNA fragments from a whole-genome library to complementary sequences of biotin-labeled probes, with secondary capture of all probe:library complexes, and post-capture amplification, quantitation, and sequencing.37
In particular, immune sequencing evaluates the adaptive immune system by assessing both the B-cell receptor and T-cell receptor (TCR) repertoire in parallel, using high throughput PCR-based deep sequencing methods.35,36,38 Immunosequencing assays may be conducted for TCR diversity assessments using forward primers for each V-gene segment and reverse primers for each J-gene segment, as well as for TCR clonality assessment of the rearranged beta chain variable regions.38 This allows quantification and identification of every B- and/or T-cell in a sample with a high degree of sensitivity and reproducibility.35 Immunosequencing may be used to potentially identify biomarkers in both peripheral blood and tumor tissue and help understand the mechanisms of action and differences among different immune therapies.36,38
In addition, whole exome sequencing, which captures all of the known coding exons in a genome, can be used to reveal the mutational load or genetic landscape of individual tumors, including nucleotide substitutions, structural rearrangements, and copy number alterations.36,37 This assay has been used to demonstrate that immune checkpoint blockade therapy specifically targets tumor-specific mutant antigens, and to identify neoantigen-specific CD8+ T-cells in patients treated with checkpoint-targeting therapies. Using exome sequencing, epitope prediction algorithms, and the tandem minigene library, both MHC class I and II neoepitopes have been identified.
MSI/dMMR status can be assessed by multiple approaches.39 It is typically detected by amplification of known microsatellite regions by PCR and quantification of the length of PCR products by gel or capillary electrophoresis; status is then determined by comparison of sample microsatellite length to normal DNA. Alternatively, MSI status can also be detected with NGS or real-time quantitative PCR melting point analysis with sequence-specific fluorescent-labeled hybridization probes. Moreover, the expression profile of mutant MMR proteins such as MutS homologs and MutL homologs can be determined by IHC using mutation-specific monoclonal antibodies.
Immunoproteomics is the large-scale study of the structure, interactions, and functions of immune-related proteins.36 Functional protein microarrays allow the simultaneous assessment of the serologic response of a large proportion of the human proteome (seromics), with sensitivity and specificity comparable to the standard enzyme-linked immunosorbent assay.36,40 Using protein microarrays, enhanced antibody responses to a higher number of antigens were detected in clinical responders to CTLA-4 inhibitor treatment than nonresponders in patients with advanced prostate cancer.41
Flow cytometry and mass cytometry platforms allow for the identification as well as functional and phenotypic analyses of subpopulations of immune cells, providing a signature of immune responsiveness. Recent efforts in flow cytometry have focused on standardizing its use and incorporating newer fluorochromes to expand its clinical application. In parallel, mass cytometry has become a platform for high-dimensional single-cell analysis, using probes labeled with heavy metal ions instead of fluorescent probes, and allowing for the simultaneous detection of many parameters.36
Multiplex IHC staining with a panel of specific antibodies offers insights into the spatial localization and distribution of structural and functional biomarkers within the TME. Cell-based assays such as the three-dimensional (3D) cell culture model represent a promising ex vivo approach for defining and analyzing immune-based biomarkers. The 3D model measures immune cell function more reliably and allows the use of primary tumor cells and autologous immune cells from individual patients.36
Challenges to Identification and Clinical Use of Immune Biomarkers
There are several practical and technical hurdles to the identification and development of biomarkers clinically relevant to immune checkpoint therapy.35,36 As alluded to earlier, there are biologic limitations to the detection of immune biomarkers in tumor biopsies. Immune biomarkers such as the PD-L1 gene are not typically mutated or amplified but show dynamic protein expression changes that are affected by location and time, as well as concurrent or prior treatments.15,31 Therefore, binary discrimination of responsiveness may not be feasible, with some data suggesting the need for more “continuous” measures. In addition, given the complexity of immune responses, they are likely not characterized by a single alteration; therefore, assaying a panel of biomarkers may be needed.15 Although reasonable concordance with samples measured across time and location has been reported, there is concern that sampling error could result in false-negative tests. Moreover, identification of immunotherapy biomarkers has been hampered by a paucity of data from large prospective trials that can adequately validate their use.3,5
Another important challenge relates to difficulties in standardization, measurement, and interpretation of biomarker assays, which are exemplified with the PD-L1 tests.5 Quantitative cutoffs for positive and negative expression of PD-L1 (1%, 5%, 10%) are not standardized and have been variably described in clinical studies across tumor types.5 Variability in the types of cells included within the scoring system has also been documented, with some restricted to only tumor staining of PD-L1, others including tumor and immune cells, and yet others including staining of only immune-infiltrating cells.5,38 Moreover, considerable variability in staining intensity and patterns have been reported with the available anti–PD-L1 antibodies used in the assays.5
Other assay features pertaining to tissue fixation, storage, and antigen retrieval are also not standardized, and therefore can result in PD-L1 degradation.31 In general, obtaining patient samples of peripheral blood, tumor cells, and TILs for biomarker analyses may pose a practical challenge, particularly since they need to be collected before, during, and after immunotherapy. Other considerations are the quality and quantity of tissue used for biomarker analyses to obtain reliable results. Although fresh tumor tissue is preferred over frozen tissue, getting access to the required amount of fresh tissue may be an issue.3,31,35
Accessibility to tests that are specific for immune biomarkers may be limited. Although some methods such as cytometry, IHC staining, and gene microarrays may be more readily accessible, access to novel technologies that are at earlier stages of development may be an issue. Also, until these tests are optimized and more routinely adopted, the costs associated with immune biomarker tests may be prohibitive.
Clinical Evidence of Biomarker-Driven Studies
There are limited clinical data available relating to biomarker-driven clinical trials that support the clinical utility of biomarkers to guide treatment with cancer immunotherapies across tumor types. The KEYNOTE-001 trial demonstrated that PD-L1 expression in at least 50% of tumor cells correlated with improved efficacy with pembrolizumab in patients with advanced NSCLC.17 This led to its current approval for the second-line treatment of advanced NSCLC in patients who are PD-L1–positive, as assessed by the companion diagnostic PD-L1 IHC 22C3 pharmDx test.42,43 However, the utility of PD-L1 expression as a predictive marker of response to nivolumab treatment in NSCLC is presently unclear, with data indicating that some patients with PD-L1–negative disease also derive benefit from its administration.29
In the phase 3 CheckMate-017 study comparing nivolumab with docetaxel in patients with previously treated squamous-cell NSCLC, PD-L1 expression was not predictive of overall response, PFS, or overall survival (OS) across the prespecified expression levels of 1%, 5%, and 10%.18 In contrast, in the parallel phase 3 CheckMate-057 study comparing nivolumab with docetaxel in patients with previously treated nonsquamous NSCLC, there was a strong predictive association between PD-L1 expression (1%, 5%, and 10%) and overall response, PFS, and OS.19 Based on these results, the FDA does not require PD-L1 testing for use of nivolumab in patients with NSCLC. More recently, results from 5 single-arm, biomarker-driven clinical trials showed that PD-1 blockade with pembrolizumab was efficacious in patients with advanced dMMR cancers including CRC, as assessed by PCR or IHC.44 Based on these data, pembrolizumab also received approval for adult and pediatric patients with unresectable/metastatic MSI-H/dMMR solid tumors that have progressed following prior treatment and without satisfactory alternative treatment modalities, or CRC that has progressed following treatment with a fluoropyrimidine, oxaliplatin, and irinotecan.45 MSI-H or dMMR tumor status was assessed using local or central, laboratory-developed, investigational PCR tests for MSI-H status and IHC tests for dMMR.
Emerging results from the proof-of-principle PANACEA trial that evaluated the combination of trastuzumab and pembrolizumab in patients with advanced HER2-positive breast cancer, who had progressed on prior trastuzumab-based therapies, demonstrated that the PD-L1–positive subgroup with 5% or more TILs present in the metastatic lesion achieved a disease control rate of 47%. None of the patients in the PD-L1–negative cohort responded to the combination therapy.22
Conclusions
Despite the recent progress made with immune checkpoint inhibitors as a new treatment paradigm in several cancer types, many challenges remain. These include properly identifying patients who will respond to cancer immunotherapy, dealing with irAEs, and addressing resistance to immunotherapy. These challenges underscore the need for validated biomarkers specific for immunotherapy. Putative biomarkers of interest include PD-1 and PD-L1 expression, TMB, MSI, dMMR, and tumor-infiltrating immune cells. However, not all of these have been fully validated, owing in part to the complex and dynamic nature of tumor–immune system interactions, as well as challenges in optimization and standardization of biomarker assays. Validation of the biomarkers in larger randomized, clinical studies are warranted to determine their true predictive value, thus paving the way for adoption of precision medicine in oncology.
References
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource [Internet]. Silver Spring (MD): Food and Drug Administration (US); 2016. Co-published by National Institutes of Health (US), Bethesda (MD). www.ncbi.nlm.nih.gov/books/NBK326791/. Updated May 2, 2018. Accessed May 4, 2018.
- Ballman KV. Biomarker: predictive or prognostic? J Clin Oncol. 2015;33:3968-3971.
- Henry NL, Hayes DF. Cancer biomarkers. Mol Oncol. 2012;6:140-146.
- Gnjatic S, Bronte V, Brunet LR, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer. 2017;5:44.
- Spencer KR, Wang J, Silk AW, et al. Biomarkers for immunotherapy: current developments and challenges. Am Soc Clin Oncol Educ Book. 2016;35:e493-e503.
- Baselga J, Cortés J, Im SA, et al. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J Clin Oncol. 2014;32:3753-3761.
- Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16:413-428.
- Hurvitz SA, Gelmon KA, Tolaney SM. Optimal management of early and advanced HER2 breast cancer. Am Soc Clin Oncol Educ Book. 2017;37:76-92.
- Gainor JF, Longo DL, Chabner BA. Pharmacodynamic biomarkers: falling short of the mark? Clin Cancer Res. 2014;20:2587-2594.
- Innocenti F, Ratain MJ. Pharmacogenetics of irinotecan: clinical perspectives on the utility of genotyping. Pharmacogenomics. 2006;7:1211-1221.
- Subudhi SK, Aparicio A, Gao J, et al. Clonal expansion of CD8 T cells in the systemic circulation precedes development of ipilimumab-induced toxicities. Proc Natl Acad Sci U S A. 2016;113:11919-11924.
- Das R, Bar N, Ferreira M, et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J Clin Invest. 2018;128:715-720.
- Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-5327.
- Nelson D, Fisher S, Robinson B. The ‘‘Trojan Horse’’ approach to tumor immunotherapy: targeting the tumor microenvironment. J Immunol Res. 2014;2014:789069.
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56-61.
- Blank CU, Haanen JB, Ribas A, Schumacher TN. The “cancer immunogram.” Science. 2016;352:658-660.
- Garon EB, Rizvi NA, Hui R, et al; KEYNOTE-001 Investigators. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018-2028.
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123-135.
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. N Engl J Med. 2015;373:1627-1639.
- Hamid O, Schmidt H, Nissan A, et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med. 2011;9:204.
- Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568-571.
- Loi S, Giobbe-Hurder A, Gombos A, et al. Phase Ib/II study evaluating safety and efficacy of pembrolizumab and trastuzumab in patients with trastuzumab-resistant HER2-positive metastatic breast cancer: results from the PANACEA (IBCSG 45-13/KEYNOTE-014) study. Presented at: San Antonio Breast Cancer Symposium; December 5-9, 2017; San Antonio, TX. Abstract GS2-06.
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207-211.
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124-128.
- Pivotal phase 3 CheckMate-227 study demonstrates superior progression-free survival (PFS) with the Opdivo plus Yervoy combination versus chemotherapy in first-line non-small cell lung cancer (NSCLC) patients with high tumor mutation burden (TMB) [press release]. Princeton, NJ: Bristol-Myers Squibb; February 5, 2018. https://news.bms.com/press-release/bms/pivotal-phase-3-checkmate-227-study-demonstrates-superior-progression-free-surviva. Accessed February 2, 2018.
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43-51.
- Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509-2520.
- Friedman CF, Postow MA. Emerging tissue and blood-based biomarkers that may predict response to immune checkpoint inhibition. Curr Oncol Rep. 2016;18:21.
- Shukuya T, Carbone DP. Predictive markers for the efficacy of anti-PD-1/PD-L1 antibodies in lung cancer. J Thorac Oncol. 2016;11:976-988.
- Manson G, Norwood J, Marabelle A, et al. Biomarkers associated with checkpoint inhibitors. Ann Oncol. 2016;27:1199-1206.
- Grigg C, Rizvi NA. PD-L1 biomarker testing for non-small cell lung cancer: truth or fiction? J Immunother Cancer. 2016;4:48.
- Vranic S. Microsatellite instability status predicts response to anti-PD-1/PD-L1 therapy regardless the histotype: a comment on recent advances. Bosn J Basic Med Sci. 2017;17:274-275.
- Chae YK, Arya A, Chiec L, et al. Challenges and future of biomarker tests in the era of precision oncology: can we rely on immunohistochemistry (IHC) or fluorescence in situ hybridization (FISH) to select the optimal patients for matched therapy? Oncotarget. 2017;8:100863-100898.
- Garibyan L, Avashia N. Research techniques made simple: polymerase chain reaction (PCR). J Invest Dermatol. 2013;133:e6.
- Gulley JL, Berzofsky JA, Butler MO, et al. Immunotherapy biomarkers 2016: overcoming the barriers. J Immunother Cancer. 2017;5:29.
- Yuan J, Hegde PS, Clynes R, et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J Immunother Cancer. 2016;4:3.
- Koboldt DC, Steinberg KM, Larson DE, et al. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27-38.
- Weber JS. Biomarkers for checkpoint inhibition. Am Soc Clin Oncol Educ Book. 2017;37:205-209.
- Menyhárt O, Harami-Papp H, Sukumar S, et al. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochim Biophys Acta. 2016;1866:300-319.
- Gnjatic S, Ritter E, Buchler MW, et al. Seromic profiling of ovarian and pancreatic cancer. Proc Natl Acad Sci U S A. 2010;107:5088-5093.
- Kwek SS, Dao V, Roy R, et al. Diversity of antigen-specific responses induced in vivo with CTLA-4 blockade in prostate cancer patients. J Immunol. 2012;189:3759-3766.
- FDA Approved Drugs. Pembrolizumab (KEYTRUDA) Checkpoint Inhibitor. www.fda.gov/drugs/informationondrugs/approveddrugs/ucm526430.htm. Accessed February 2, 2018.
- Agilent Pathology Solutions. PD-L1 IHC 22C3 pharmDx Interpretation Manual. Santa Clara, CA. www.agilent.com/cs/library/usermanuals/public/29158_pd-l1-ihc-22C3-pharmdx-nsclc-interpretation-manual.pdf. Accessed May 7, 2018.
- Diaz L, Marabelle A, Kim TW, et al. Efficacy of pembrolizumab in phase 2 KEYNOTE-164 and KEYNOTE-158 studies of microsatellite instability-high cancers. Presented at: European Society for Medical Oncology Congress; September 8-12, 2017; Madrid, Spain. Abstract 386P.
- FDA approves first cancer treatment for any solid tumor with a specific genetic feature [press release]. Silver Spring, MD: US Food and Drug Administration; May 23, 2017. www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm. Accessed February 2, 2018.